One of the main reasons that inbreeding is harmful is that it increases the level of homozygosity in the genome, which in turn exposes the organism to the effects of deleterious recessive variants (the Habsburgs are a classic case of this happening in humans).
However, inbreeding can also help purge harmful mutations by allowing selection to act against mutations that it was previously “blind” to when they were in a heterozygous state (assuming that most such mutations are mostly or entirely recessive).
This week in Nature Plants, there is new paper looking at the effects of purging in some samples of maize that were forced to undergo selfing for several (6 or more) generations.
The genome-wide dynamics of purging during selfing in maize
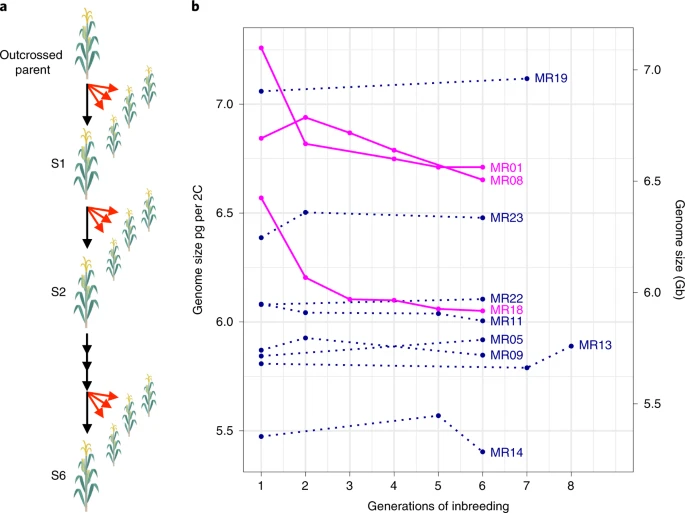
I want to draw your attention to one particularly interesting effect of purging that the researchers found: 3 of the 11 maize lines saw a dramatic decrease in genome size in only a few generations:
a , Schematic of the study design. An outcrossing parent was selfed to make the S1 generation and then subsequently selfed until S6 or higher. The selfed, single-seed descent lineages are represented by black arrows. Our study used sibling seed sampled from each generation, represented by red arrows. b , Estimates of genome size, in pg per 2C content, across generations of selfing. Each of the 11 lines is represented. Dark lines represent significant decreases of GS. Dotted lines show no detectable changes in GS over time. Sample size varied between one and three for each line and generation (see Supplementary Table 1 for details) with a total of n = 96 plants sampled. See Supplementary Table 2 for raw values, Supplementary Tables 3 and 4 for statistics, and Supplementary Fig. 3 for a detailed plot of the raw data per line.
This represents a loss of about 200 - 500 MB in these three lines in a very short amount of time, which is quite amazing when you stop and think about it! Notably, most of what was lost appears to have been transposable element sequences, which are both extremely common in maize and also prone to being deleterious.
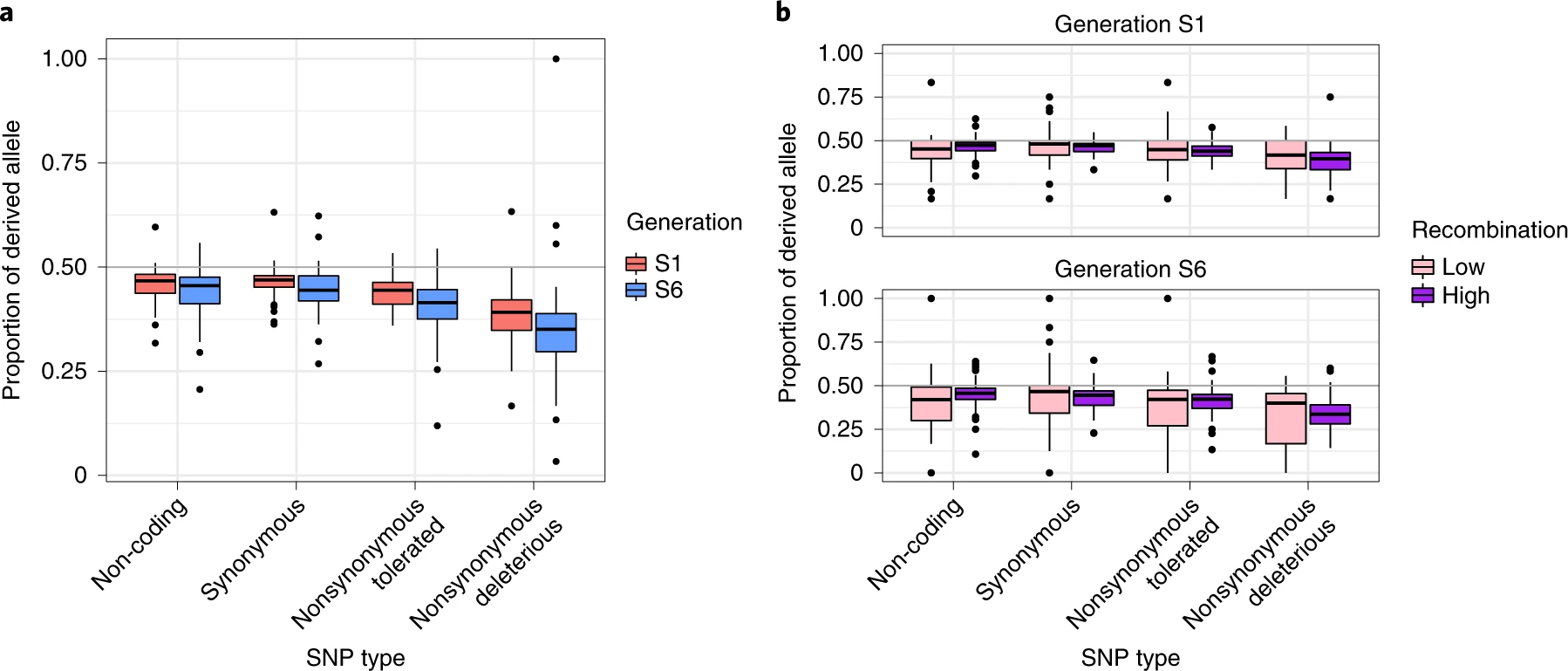
Consistent with the fact that selfing promotes purging, the researchers also found that putatively deleterious single nucleotide variants (SNVs) were removed at a higher rate than SNVs with little predicted functional effect. This was especially true in regions of higher recombination, where selection is more efficient:
a , The proportion of the derived allele for the four mutational classes predicted by SIFT—that is, non-coding, synonymous, nonsynonymous tolerated and nonsynonymous deleterious. The graph reports the proportion for generations S1 and S6 across six lines (MR01, MR08, MR09, MR18, MR19 and MR22). P d was averaged across individuals for each of the ten chromosomes and six lines separately ( n = 60 biologically independent chromosomes for each bar of the plot, n = 480 total samples across generations and categories). b , As in a , except the genome was separated into high- and low-recombination quartiles, illustrating that purging occurs more rapidly in high-recombination regions. As in a , n = 60 for each bar of the plot. Box plot as described in Fig. 2.
Unfortunately, the paper is paywalled, but I think it’s really nice work and wanted to highlight some interesting points.