So, I recently had another conversation with @anon68328710 on her channel. We did jump around different things and mostly still disagreed, but I did think it was a very nice talk. Just to briefly summarize the points we discussed if anyone is interested to watch the whole conversation.
I also have to apologize upfront. I promised Rebekah that I would be brief here regarding the topic of Alternative transcripts, but I just couldn’t help myself. Rebekah is free to skip to point 5 about alternative transcripts.
1. Metaphors
The first thing we talked about was the issue of ‘intentional’ language in biology; e.g. why do we say a protein or RNP (e.g. spliceosome) is “recognizing” a splicing site at the introns, as if there is a mind involved. She was taking all of this as an indication that all of this is designed. However, I think the issue is that such figurative language is very pervasive in general. I gave example of chemical properties being described as ‘hydrophobic’ or ‘hydrophilic’ and the fact that magnets are ‘attracted’ to each other. Of course, no chemical is ‘afraid of water’ and magnets are not falling in love, but in most instances we don’t notice such anthropomorphic or metaphorical notions. Such examples is pervasive in language in general. Many words originated via semantic shifts sometimes first involving metaphorical usage, but then this new usage became custom (via quotidianisation), and the original meaning was lost resulting in a dead metaphor; e.g. broadcast.
So to me, this seems more of a quirk of language that we have inherited and are stuck with rather than an indication that biology is designed. That’s not to say that metaphorical language can not be misleading and is not an issue to be addressed (see good reviews here and here). I have repeated the criticisms of Daniel Nicholson regarding ‘machine analogies’ and have discussed that on this forum before. I also recommend Susan Oyama’s book ‘The Ontogony of Information’ which goes deeply into this subject, particularly in chapter 5 regarding what she refers to as the ‘Homunculoid Gene’.
2. Abiogenesis
We also talked a bit abiogenesis. We also briefly mentioned Dr. James Tour and his debacle with Professor Dave, but I don’t want to reiterate that disaster here. I made an off the cuff statement about how the origins of life is… at least according to some (e.g. Eric Smith and Nick Lane)… is not an accident of but a law-like consequence of relieving an environmental stress like a ball rolling down a hill or a lightning bolt. What happens on a planet like the earth is that the rocky interior is ‘reducing’ (rich in electron donors such as iron) while the atmosphere interacting with solar radiation becomes more ‘oxidizing’ (rich in electron acceptors). That turns the planet into a giant battery, possessing a stress (or disequilibrium) of redox chemistry which is focused at places where new rock interacts with the aquatic exterior. Specifically, when electron donors produced from the interior (H2) interacts with electron acceptors (CO2) they “want” to react to release the energy (forming CH4). However, this reaction is frustrated by kinetic barriers, but the way that this is facilitated through biochemistry appears to be paths of least resistance; analogous to how a lightning bolt emerges since it is the path of least resistance that releases the build-up charges in thunderstorms. That does not make a scientific hypothesis, but I do find it intriguing. Especially since one of the implications is that ‘life’ should be properly understood as a planetary phenomenon, at the level of the biosphere instead of an individual organism.
A few recommendations: watch this talk by Eric Smith. The book he co-authored with the late Harold Morowitz is also a MASSIVE 600+ page beast to read… but be warned… it’s not a light read for laymen. This pop-sci article is much better for beginners. Lastly, I would also gladly recommend looking up Nick Lane and his books. While the aforementioned book by Smith and Morowitz is by far the richest book I have ever seen on this subject, Nick Lane’s style of writing is much more entertaining and easier to digest for people who don’t have a lot of science background.
3. Science
This was an issue that we repeatedly fell back onto at different points in time. One of my main sticking points with ID-Creationism is the lack of what they bring to the table. Most of their arguments is negative; e.g. evolution cannot explain [insert complex structure here]. While one can argue endlessly over whether any hypothesized scenario to explain, for example, the spliceosome (more on that later) is accurate or not; at the very least, there is something to discuss on that side. ID-Creationism does not provide anything to explain the spliceosome… specifically in the sense that the hypothesis is testable. Everything deemed unexplainable by naturalistic means is considered an unexplainable miracle which are untestable. One is free to believe that, but to me that would get us stuck in a corner and science can’t move forward regarding that issue. Que the famous Sidney Harris’ cartoon. Additionally, I have made clear that I do not believe that if we found a complete explanation for the spliceosome or the origin of life would be incompatible with theism. I don’t see why theist could not look at his and come a way thinking that those are the miracles that God performed, and it’s amazing that science was able to figure out how He did it. The problem I have is with the type of creationism that proposes inexplicable miracles which are science stoppers.
After I gave such an answer, Rebekah would ask me at what point I would consider such inexplicable miracles. For how long does science have to fail at explaining something until we throw our hands up and give up. I would say at no point… because at no point are you able to differentiate between something that can be explained but we haven’t yet done so, and something that is unexplainable. Arguing that the former implies the latter will always be a negative argument that appeals to our ignorance.
4. Evolution of the spliceosome
Rebekah started the stream showing a review paper that discusses the origin of the spliceosome. This is one criticism that I have made against Rebekah. When she is reviewing a paper like this, she highlights parts that can be summarized as the conclusions made by the author. Of course, Rebekah says she disagrees with said conclusions, but my main issue is that she tends to skip the parts where they discuss the evidences in support of the conclusions.
For example, the first paragraph Rebekah highlight is where the authors briefly describe the generally accepted hypothesis that the spliceosomes in eukaryotes evolved from self-splicing group II introns found in bacteria. The paragraph also happens to have no citations. It’s just a general description outlining the mainstream hypothesis. Rebekah rails against this paragraph as ‘just evolutionary story telling with no evidence’… however…. quoted below is the very next paragraph, which provides many details and references in support of the hypothesis. This is not the only paragraph that does this. But Rebekah never shows this or any similar paragraph, much less than discussing it. She moves on to another section much further down into the paper.
Structural and functional evidence suggests that spliceosomal and group II self-splicing introns are evolutionarily related. Both types of introns are spliced through a similar two-step catalytic reaction that relies on an endogenous adenosine (the BP), and releases the excised intron as a lariat structure. The two intron types have similar boundary sequences (GT-AY in group II introns and usually GT-AG in spliceosomal introns, although some U12 introns are AT-AC) (Lambowitz and Zimmerly 2011), and there are striking structural similarities between key regions of group II intron domains and spliceosomal snRNAs (Lambowitz and Zimmerly 2011). These include at least (i) domain DV and U6 snRNA, with divalent metal-ion binding sites involved in catalysis and similar base-pairing interactions (Jarrell et al. 1988; Peebles et al. 1995; Yu et al. 1995; Abramovitz et al. 1996; Konforti et al. 1998; Yean et al. 2000; Shukla and Padgett 2002), further supported by crystal structure (Toor et al. 2008; Keating et al. 2010); (ii) ID3 subdomain and d–d′ motifs and U5 snRNA stem loop, involved in the recognition of 5′ and 3′ exons (Hetzer et al. 1997); and (iii) DVI and the U2-intron pairing that include the BP adenosine (Schmelzer and Schweyen 1986; Parker et al. 1987; Li et al. 2011). In addition, it has also been shown that extracts of snRNAs can catalyze both splicing reactions without proteins in vitro (Valadkhan et al. 2007, 2009), in a similar manner to complete group II introns.
I find such instances problematic. When I point this out, Rebekah says she tends to focus on the highlights of the paper in order to maintain the attention of the audience. I don’t find that a sufficient reason to skip key sections such as the one I quoted above. So, right now Rebekah tends to review a paper in a way that skips over the parts that discusses the evidence. I would rather see her do the following: Show what the authors hypothesize, and then explain (the main) the reasons or purported lines of evidence they give in support of their conclusions, and then she gives her opinion on this. Even if Rebekah still thinks that the evidence provided did not convince her, at the very least this would be a big improvement from skipping over these parts entirely.
Just a few additional notes on this. I found a more recent (2017) review paper and a very recent (2025) research paper on this subject. I would like Rebekah to read the one from 2017 since that includes some critical commentary from reviewers, especially from Ford W. Doolittle and Eugene V. Koonin. I think that gives a good example of how peer review is (should be) done and perhaps would dispel her idea that “things are just assumed” when it comes to peer review of these papers.
5. Alternative transcripts
This is the point that I wanted to write specifically on this forum. I promised Rebekah to give more details about alternative transcripts, which we disagree on. Specifically, during our talk we noted that the human genome has 20,000 genes, but Rebekah says we have between a hundred thousand to a million different proteins, and those facts point to alternative transcript being functional. I disagree. I think there is good reasons to conclude that most alternative transcripts that we detect are errors of splicing, and these transcripts are cleaned up by processes such as nonsense-mediated decay. There are examples of alternative splicing where each of the alternative transcripts have been shown to be functional, but these are a minority of the genes.I touched on this during the conversation, but I can give more details here. I will also be borrowing points made by Larry Moran from his book ‘What’s in your genome’.
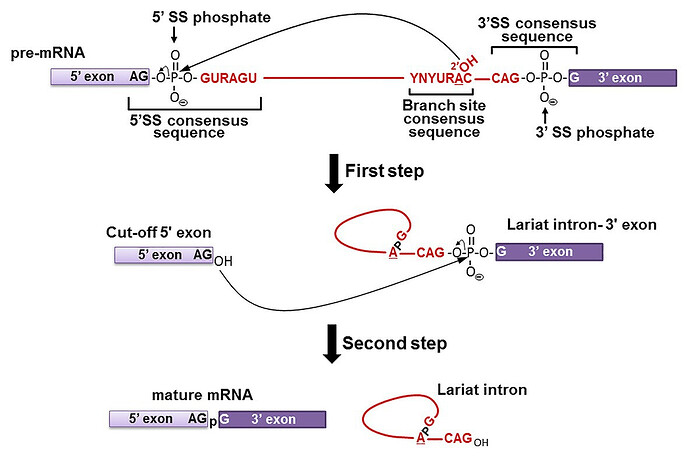
The spliceosome recognizes sequences that are present at the 5’ and the 3’ end of the introns. There is also a sequence in the middle that is called the ‘branch point’. The first step involves the 2’ OH group of an Adinosine nucleotide at the branch point reacting with oxygen in the 5’-3’ phosphodiester bond at the 5’ end of the intron. This results in the release of the 5’ exon, and producing an very unusual 2’-5’ phosphodiester bond. Next the 3’OH of the 5’ exon does pretty much the same reaction at the 5’-3’ phosphodiester bond at the 3’ side. This ligates the exons together, and releases the intron ‘lariat’.
The specificity of the spliceosome is mediated by the sequences at these points splicing sites. The spliceosomal proteins contains short pieces of RNA (snRNA) which are able to bind to the splicing sites in the mRNA via complementary base pairing; e.g. U1 pairing at the 5’ splice site and U2 at the branch point site.
The degree of complementarity between these determines the strength of the hydrogen bonding that can be achieved. If the complementarity is insufficient then the bonding strength is too weak and the spliceosome won’t cut the sites. That’s how the spliceosome is able to “recognize” the splicing sites (it’s not a cognitive or mental). Now, the key thing is that the degree of complementarity. Some sequences are more easily “recognized” by the spliceosome. The ideal sequences that the spliceosome “recognizes” results in a splicing accuracy of ~99.999%. There is a small degree of error because the molecular world is inherently noisy, although by the standards of DNA replication and transcription this error rate is actually relatively high. Furthermore, most splicing sites found in our introns do not have this ideal sequence. They may deviate from it by 1 or 2 nucleotides. This could decrease the accuracy down to 99.9% and this will produce transcripts 0.1% of which will be splicing errors. We are able to detect these transcripts because RNA sequencing is very sensitive (an RNA sample can contain hundreds or thousands of mRNA copies per gene). This is also consistent with the fact that, with few exceptions, the vast majority of alternative transcripts do not produce a polypeptide.
When Rebekah mentions that we have much more functional proteins compared to genes, I think she is referring to post-translational modifications (e.g. phosphorylation, glucosylations, and several hundred other types). This does not correspond to alternative splicing (that’s pre-translation). Also, we should take care to avoid concluding there such post-translational modifications means that each gene produces dozens of proteins. Larry Moran has also made remarks on that as well.