Life is younger than we suppose, and it never evolved.
The human genome isn’t mostly functional with respect to sequence specific function, so evolution isn’t wrong.
So how does he manage to hold that view, given the abundant evidence that everything is not going extinct, and has not done so? Further, what we see from his “work” is that either everything is going extinct or there’s something wrong with his model. He makes one suggestion, that there’s some mysterious force preventing extinction, but the simpler hypothesis is that his parameters and model do not reflect reality.
How is this at all relevant? It feels like you’re just squirting out as much ink as possible, trying to confuse the thread.
The movement of DNA is what moves transcription factors through itself, so we have such large genomes to help transcription factors diffuse, something they would otherwise have trouble doing through such a… large genome? What point as you trying to make here?
Sal, you’ve had enough scientific training and experience in rhetoric that I’d like to think you’d understand that when you’re making an argument, you don’t just throw out random citations and quotes - you’re supposed to explain how they’re relevant to you specific point!
That’s a hypothesis completely at odds with all other data. Try again.
Even in closely related that creationists would call members of the same “kind”, there is obviously no selection pressure acting to preserve the sequences that you claim are functional as redundancies.
Maybe you need to think that DNA is polyfunctional in other ways. Sequences can affect resonant vibration and if the model is correct will affect Transcription Factor transport. You’re being waaay to quick to assume DNA is 90% functionless. I’m giving you reasons to reconsider.
If you demonstrated that mutations to non-coding DNA changed the “resonant vibration” of DNA in such a way that inhibited TF migration, that would be a start, but then you’d also have to explain why we see no selection against such mutations.
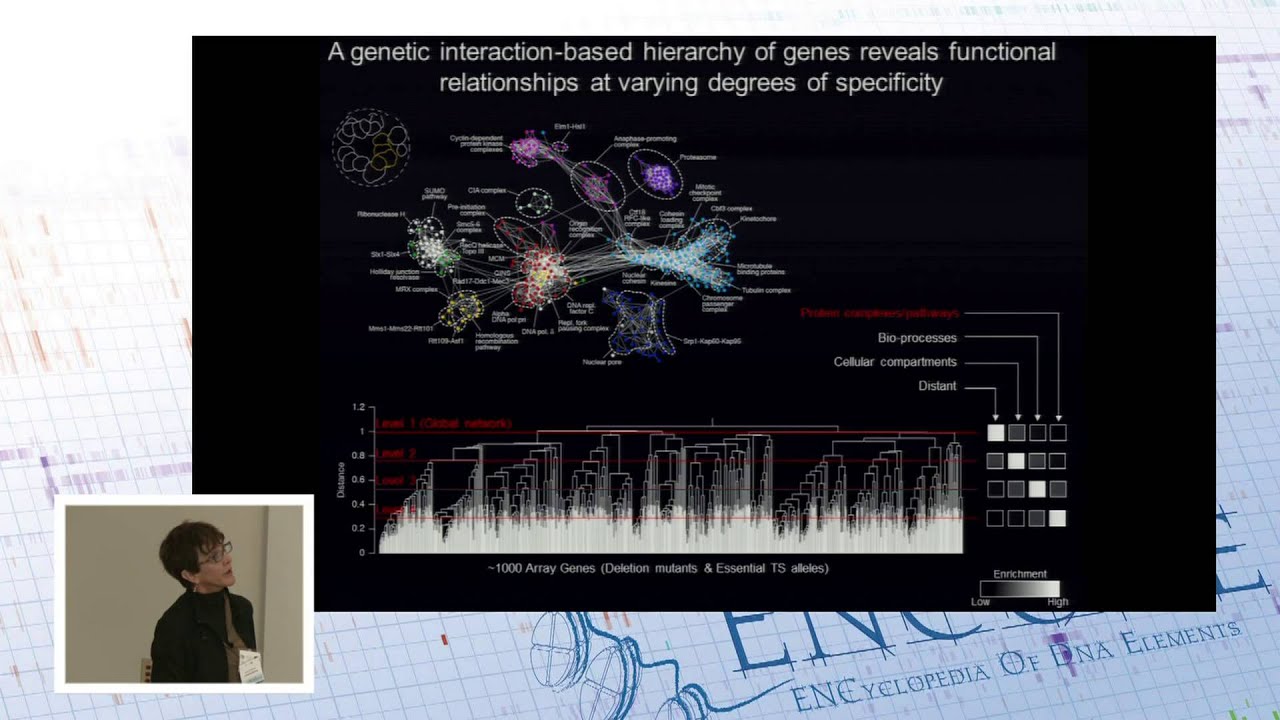
Because selection can’t see all functional compromise, I gave an example with D4Z4 repeats and more to the point Brenda Andrews gave a presentation at the ENCODE 2015 why redundancy or robustness can’t be seen by fitness tests easily. She gave evidence of the problem based on her single vs. double knockout high throughput experiments on yeast genomes.
See here:
But in anycase, DNA folding is affected by sequence, so don’t dismiss that one can freely mutate DNA and it won’t affect it’s 4D behavior. Here is evidence of that from the study of DNA Origami in 2019:
It isn’t an assumption. It is a conclusion based on the rate of accumulation of mutations in those sections of DNA. 90% of the human genome bears strong evidence for a lack of selection against deleterious mutations. Again, we are following positive evidence, not just assuming.
Why are you overlooking the 3rd possibility, ie., the idea that a yet unknown natural force or mechanism preserves species from GE?
And to set the record straight, Sanford’s thesis is not what you say (you’re building a straw man, again). It is more as follows: If the neo -Darwinian theory is true, then all species must go extinct. It follows that if it is not the case that species go extinct, then neo-Darwinian theory is false and something else must be at work that account for species resistance to GE.
Because selection can’t see all functional compromise, I gave an example with D4Z4 repeats and more to the point Brenda Andrews gave a presentation at the ENCODE 2015 why redundancy or robustness can’t be seen by fitness tests easily. She gave evidence of the problem based on her single vs. double knockout high throughput experiments on yeast genomes.
Recessive deleterious alleles would be under selective pressure. Natural selection can see them.
But in anycase, DNA folding is affected by sequence, so don’t dismiss that one can freely mutate DNA and it won’t affect it’s 4D behavior. Here is evidence of that from the study of DNA Origami in 2019:
There’s a huge difference between designed DNA origami on the scale of tens or even a few thousand bp versus megabase-scale TADs and other regulatory structures.
Why are you overlooking the 3rd possibility, ie., the idea that a yet unknown natural force or mechanism preserves species from GE?
I wouldn’t call it “overlooking” when we ignore stuff that is made up on the spot and is lacking evidence.
It is more as follows: If the neo -Darwinian theory is true, then all species must go extinct.
That is false. His entire thesis depends on his own claim that most mutations are slightly deleterious. He has yet to support that claim.
It isn’t an assumption. It is a conclusion based on the rate of accumulation of mutations in those sections of DNA. 90% of the human genome bears strong evidence for a lack of selection against deleterious mutations. Again, we are following positive evidence, not just assuming.
That is unfortunately circular reasoning, the same sort of reasoning that said ERVs are junk, and now we suspect many of them are important targets for a variety of molecular machines.
When I saw claims of co-evolution of ERVs with KRAB Zinc Finger proteins, it became evident there was lots of circular reasoning since the co-evolution of even modest Zinc Finger arrays is absurdly improbable.
That is unfortunately circular reasoning, the same sort of reasoning that said ERVs are junk, and now we suspect many of them are important targets for a variety of molecular machines.
What is that circular reasoning?
How do you explain the difference in sequence conservation between exons and introns?
When I saw claims of co-evolution of ERVs with KRAB Zinc Finger proteins, it became evident there was lots of circular reasoning since the co-evolution of even modest Zinc Finger arrays is absurdly improbable.
Which ERV’s? All of them, or just a few of them?
If a deleterious mutation happens in one viral genome, how are the descendants of that virus able to outcompete the viruses without that mutation?
A large part of the answer has to do with the phenomenon of bottelnecking.
A large part of the answer has to do with the phenomenon of bottelnecking.
References? Data?
Why are you overlooking the 3rd possibility, ie., the idea that a yet unknown natural force or mechanism preserves species from GE?
Sure, let’s just call it MAGIC. All science all the time. ![]()
If the neo -Darwinian theory is true, then all species must go extinct.
What a ridiculous non-sequitur. That conclusion does not follow from the premise. Besides we weren’t talking about species going extinct but life going extinct from GE. New species arise from older species all the time. It’s called common descent and has been happening for 3.5 billion years. Does the process of speciation somehow magically reset the genome of the new species to “perfect” so GE starts all over again? Or maybe it’s the Space Aliens with their ray gun? ![]()
Sorry, Sanford’s GE idiocy is deader than dead. It was a scientific non-starter from the beginning.
There’s a huge difference between designed DNA origami on the scale of tens or even a few thousand bp versus megabase-scale TADs and other regulatory structures.
God-made megabase-scale TADs are far more complex folding designs with architectural proteins than the puny designs of man-made DNA of only a few thousand bases.
As I pointed out earlier:
it has been proposed that interphase DNA has an irregular 10-nm nucleosome polymer structure whose folding philosophy is unknown. Nevertheless, experimental advances suggest that this irregular packing is associated with many nontrivial physical properties that are puzzling from a polymer physics point of view. Here, we show that the reconciliation of these exotic properties necessitates modularizing three-dimensional genome into tree data structures on top of, and in striking contrast to, the linear topology of DNA double helix.
So why the eagerness to make sweeping pronouncements and adevertise those pronouncements as science? Those are faith statments that DNA is 90% junk, they are not experimental statements.
All I hear so far is trivializing of the physical influence of sequence on polymer folding. That’s dismissiveness based on evolutionary thinking, not on theories of physics, chemistry, observation and experiment. Hence these pronouncements that there is no aggregate sequence specificity are pre-mature at best, pure faith statements and dogma at worst.
That is false. His entire thesis depends on his own claim that most mutations are slightly deleterious. He has yet to support that claim
No serious population geneticist would dispute the fact that most non neutral mutations are slightly deleterious. Given that most of the genome of an RNA virus such as Influenza is functional, it follows that most mutations occurring in Influenza are slightly deleterious.