The reason I’m going through all these details of introns, etc. is that Sanford’s genetic entropy thesis is supported by the spirit of this statement by Graur:
If ENCODE is right, evolution is wrong
But we can just as well say
If the percentage of human genome function is high, evolution is wrong
This can be argued in multiple ways, but the simplest is the Bonker’s formula
\large {NumberKidsPerFemale} = 2 e^U
where U is the number of mutations.
It just so happens that for every 1% of the genome that can be reasonably argued is function, that makes the problem about 5.4 times harder for evolution because each human female needs to be making 5.4 times as many kids. At one end of the extreme, even Graur said, each female would need on the order of 10^35 kids. And commented, “this is clearly BONKERS!”
Introns account for a lot of non-coding DNA. Unfortunately, the introns are not a disjoint group from other ncDNAs like ERVs and Alus since these can be inside introns…
That said, ERV were also once said to be functionless parasitic junk. It turns out they are often targets of the zinc finger proteins like the CTCF zinc finger protein. And this plays an important role in gene regulation. Here is one example:
Endogenous Retroviruses Function as Gene Expression Regulatory Elements During Mammalian Pre-implantation Embryo Development - PMC
Endogenous Retroviruses Function as Gene Expression Regulatory Elements During Mammalian Pre-implantation Embryo Development…ERVs provide binding sites for a chromatin organizer, and then participate in the formation of a high-order chromatin structure.
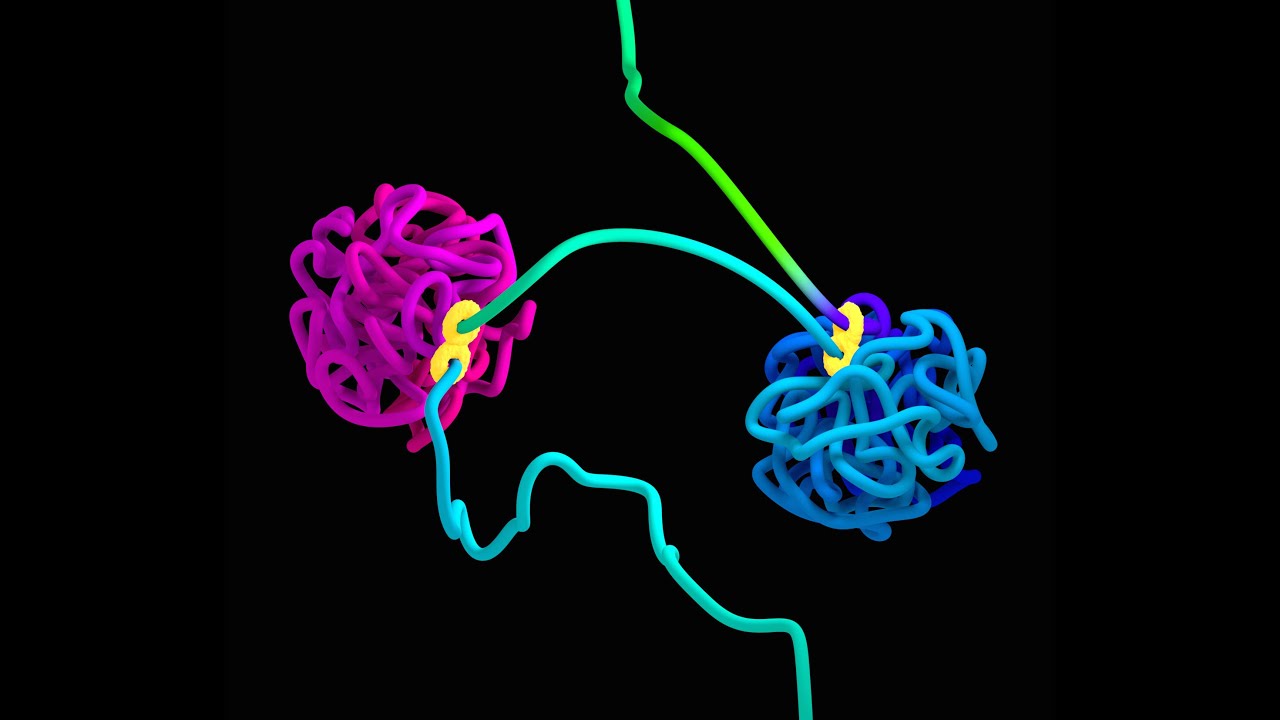
There are other examples of zinc finger proteins like KRAB-KZNF. The DNA-binding site in the diagram below is often an ERV or some other non-coding element!
But what is most amazing is how the ERVs or other CTCF binding sites have to be gramatically or geometrically oriented to enable creation of regulatory loops.
See this 3-minute video, and remember each time you see the “CTCF motif”, that is often an ERV or some other “junkDNA” that makes the system possible! Note the “junkDNA” has to be pointed in the direction of the arrows to make this system work.
This also suggests the ERVs (or other “junkDNA”) aren’t randomly positioned unless it was an accidental mutational deviation from a healthy state. Given these kinds of DNAs can jump around, such transposition mutations to the DNA may create defects that will disrupt the normal looping of DNA resulting in genetic degeneration over time.