Donald Hilvert of the Swiss Federal Institute of Technology (ETH) Zurich and his colleagues show that the this hypothesis is plausible by making it happen in the lab ( Science 2021, DOI: 10.1126/science.abg2822). “You can take a bacterial protein that has no starting affinity for nucleic acids and teach it how to bind its own encoding mRNA, package it, and protect it against nucleases,” Hilvert says.
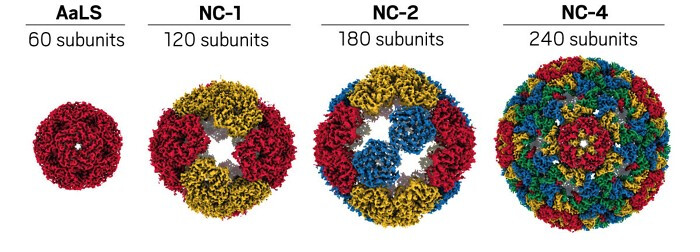
The researchers started with a bacterial enzyme called lumazine synthase, which naturally forms 60-subunit nanoscale cages. The researchers redesigned the protein by adding a peptide that tightly binds an RNA sequence called BoxB; this peptide was designed to trap the RNA the team hoped to evolve the protein to package. Meanwhile, they added BoxB to both ends of the RNA sequence encoding the protein. They then subjected the protein to multiple rounds of directed evolution in which they randomly mutated the gene encoding the protein and selected for variants that could package the target RNA and protect it from nucleic acid–cleaving enzymes.
The resulting container combined 240 protein subunits and efficiently packaged the full-length RNA. Intermediate versions had fewer subunits with large pores that let nucleic acid–cleaving enzymes in. During evolution, the protein underwent structural changes in which monomers interlaced with one another by swapping domains and forming trimeric building blocks. Those building blocks further assembled into pentamers of trimers. The final capsid consisted of 12 pentamers of trimers plus an additional 20 trimers in a spherical shape with small pores. The interlacing reduces the size of the pores compared to the original, which helps block nucleases.
The work is a “multidimensional tour de force that experimentally recapitulates processes by which viruses might have emerged from free-living organisms,” says Charles W. Carter Jr., a structural biologist at the University of North Carolina at Chapel Hill.