2 hrs above:
Ah. What an odd and apparently random assemblage of questions. They seem united only in that most of them are intended as zingers (zing-fingers?) directed at evolution and/or anything else that isn’t YEC. I do note that the nested hierarchy of life appears twice on that list; I think those may be two of the questions Sal would miss.
Spelling flame: “Olbers’”.
Well, I’ll post my answers in my course. And FWIW, I’ve written on some of this in fragmented places on the net.
For example, if we treat the NON-thermodynamics macrostate of system of fair coins as the percent of heads, but rather the information entropy of heads/tails I already provided the computation here in terms of combinatorics using C(n,r):
SSDD: a 22 sigma event is consistent with the physics of fair coins? | Uncommon Descent
But, I did provide an estimate of thermodynamics entropy of the same system in terms of molar entropy (there were some typos, but the idea is correct as affirmed by Gordon Davission):
But you just asserted that I didn’t understand the law of large numbers. But I stated textbook definitions and provided examples of what the law of large numbers predicts.
But, I should point out, you’re taking issue with ME personally, not the material that is stated.
Well, Dr. Harshman, I said the list is subject to revision. I and my students thank you for your free-of-charge editorial review to correct such things a typos and redundancies. I look forward to your invaluable future input as well on the course materials.
More to the point, it’s impossible to take this list seriously. It seems a random assemblage of stuff you happened to think of, and I doubt it could have been the result of more than 15 minutes of thought.
You could state specifically what you think I can’t answer, other than that, that’s just speculation and sounds like a total cheap shot at me personally.
Compare the total thermodynamic entropy of a live human to a frozen dead rat.
Use the law of large numbers to estimate the probability of macrostates in terms of the count of microstates.
What are the features of Eukaryotes?
List the problems in evolving Eukaryotes.
Describe the action of the HOTAIR lncRNA.
What are promiscuous domains?
Estimate the likelihood a random-frameshift mutation will create a zinc-finger protein.
Identify key residues of a metal binding protein using sequence alignments.
Well, that’s actually 8. One strike against me, I guess.
Here are some striking cirrus clouds with regular oscillations:

Do they also violate the law of large numbers because of those regular features? What is the probability of those water molecules gathering together in that exact formation?
Here is a comparable calculation using standard molar entropies of an ice cube and a living human.
The living human has far more thermodynamic entropy than a frozen dead rat.
That was done for NON-thermodynamic entropy here, which was the aim of the lesson:
There are 501 macrostate, and the number of microstates that instantiate each macrostate in this NON-thermodyanmic example is stated by C(n,r).
The good news - the general trend is correct.
The bad news - the additional constituents, and the teaching moments associated with thermodynamic considerations thereof, are completely missing.
As I was saying …
This can be done across species, but I just showed one metal binding protein. The Cysteins and Histidines, highlighted are the key residues that can be done cross-species. I took the liberty of showing how this is even evident in a single mult-zinc finger protein – zinc metal ions bound to the each of the classical C2H2 zinc fingers, where I aligned the DOMAINS in the same protein. It’s not like you haven’t seen me post it already!
This the fold with the metal binding:

I could show key residues for non-metal binding protein beta lactamase/nylonases like the one below. And again, i’s not like you haven’t seen me post on this since you commented on a paper I wrote regarding it. The key residues are Serine and Lysine and conform to x-ray crystallography experiments:

But it seems your determined to make the issue about my understanding, not about the material stated. And even then, the speculation you offered, is not consistent with the data you yourself have already seen in these discussions.
Well, again, it’s not like you haven’t seen me talk about it, like here:
Developing College-Level ID/Creation Courses - #64 by stcordova

Here is the popular version I wrote for a creationist website:
Pinpoint Navigation and Propulsion in a Seemingly Random Soup – CEH
In 2007, John Rinn discovered a lncRNA (long non-coding RNA) transcribed from DNA on human Chromosome 12 that would somehow navigate and land at a specific location on human Chromosome 2 by riding a molecular “bus” known as the Suz12 protein. It was the first example of a transcript from one chromosome influencing the expression of a gene on another chromosome. This epigenetic action, he found, was a crucial part of cell signaling for differentiating skin cells in the body. It’s why the skin cells in the sole of the foot, for instance, have different qualities than skin cells in the lid of the eye.
Amazingly, Rinn began his research suspecting all he would come up with was “hotair” rather than a real discovery. He thus named this amazing RNA molecule HOTAIR. When his research was published, the journal editors hailed it as the greatest article in their history, but the name HOTAIR was retained. It stands for ‘HOX transcript antisense RNA’.
How this feat of navigation and propulsion by the SUZ12 protein that shuttles the HOTAIR lncRNA from a specific location on Chromosome 12 to a specific location on Chromosome 2 could be accomplished in a seemingly random soup of chemicals boggles the mind. After all, there are over a billion locations in the nucleus to park HOTAIR on a particular stretch of DNA, not to mention the additional challenge that DNA is a moving target!
What Rinn’s research demonstrated is that many parts of DNA formerly thought to be junk could serve as navigational markers, like street signs as well as highways that allow molecules to sail the winds of Brownian motion and deliver molecular packages with pin-point accuracy.
It intuitively suggests a very well conceived system of highways and shuttles where DNA plays a role. We tend to think of DNA solely as a blueprint, but DNA could also serve multiple roles as a means of navigation and propulsion!
The hypothesis that DNA serves multiple roles is furthered by a recent related development regarding proteins known as transcription factors. According to Phys.org, “Rattling DNA hustles transcribers to targets” –
Here is the term paper I wrote on Chromatin modifications related to HOTAIR from which the above essay was inspired:
https://1drv.ms/w/s!AllCu5MwyX4PhlfSzJB6xFVD9wpS?e=ytjHfp
That name caught my eye. Not Sanford, Seaman. I met Josiah Seaman last year at the big evolutionary biology congress in Montpellier, and I noticed that he was a student in Richard Buggs’ group at Kew Botanical Gardens. I wondered at the time if he might have any ID sympathies like Buggs, but never really looked into it. Now I see that he co-authored his first paper with Sanford (the skittle paper above), and also published a conference paper in the “Biological Information: New Perspectives” book in 2013, both while supported by Sanford’s FMS foundation. Maybe my intuition was right.
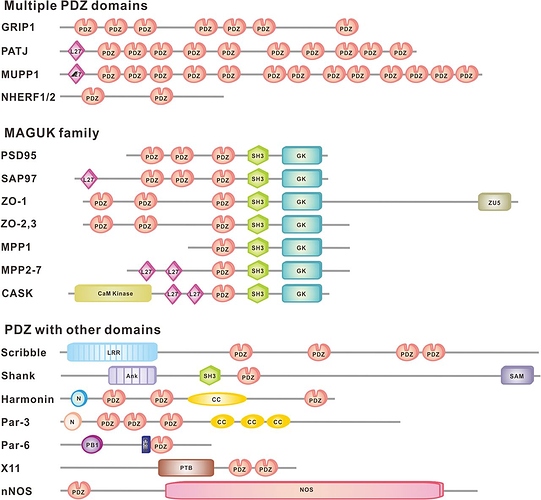
You demanded slides from my 2-hours presentation that was entitled “promiscuous domains” and I provided them. Here they are again:
But anyway the formal paper is here with Koonin as co-author:
Evolution of protein domain promiscuity in eukaryotes.
National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Maryland 20894, USA.
Numerous eukaryotic proteins contain multiple domains. Certain domains show a tendency to occur in diverse domain architectures and can be considered “promiscuous.” These promiscuous domains are, typically, involved in protein-protein interactions and play crucial roles in interaction networks, particularly those that contribute to signal transduction. A systematic comparative-genomic analysis of promiscuous domains in eukaryotes is described. Two quantitative measures of domain promiscuity are introduced and applied to the analysis of 28 genomes of diverse eukaryotes. Altogether, 215 domains are identified as strongly promiscuous. The fraction of promiscuous domains in animals is shown to be significantly greater than that in fungi or plants. Evolutionary reconstructions indicate that domain promiscuity is a volatile, relatively fast-changing feature of eukaryotic proteins, with few domains remaining promiscuous throughout the evolution of eukaryotes. Some domains appear to have attained promiscuity independently in different lineages, for example, animals and plants. It is proposed that promiscuous domains persist within a relatively small pool of evolutionarily stable domain combinations from which numerous rare architectures emerge during evolution. Domain promiscuity positively correlates with the number of experimentally detected domain interactions and with the strength of purifying selection affecting a domain. Thus, evolution of promiscuous domains seems to be constrained by the diversity of their interaction partners. The set of promiscuous domains is enriched for domains mediating protein-protein interactions that are involved in various forms of signal transduction, especially in the ubiquitin system and in chromatin. Thus, a limited repertoire of promiscuous domains makes a major contribution to the diversity and evolvability of eukaryotic proteomes and signaling networks.
Here are some pictures of domain promiscuity:

and
Asked for. Not demanded.
That’s not a very good description.
You’re welcome to offer what you think is a better description. I’m ok with that.
T_aquaticus,
Regarding the cirrus clouds, I don’t think they violate any statistical law. But whether it is designed or not designed, is not the concern of the course I’m putting together. Unlike other ID proponents, I say there are only certain limited systems that can be tractably analyzed in terms of probability.
The design argument is for systems that can be reasonably described in terms of experiments to determine what the statistical and/or chemical expectation is under various conditions.
Many abiogenesis experiments can’t even polymerize monomers in a plausible solvent (like water), much less even get to the stage where the law of large numbers can be invoked, but even when condensation reaction and/or polymerization mechanisms are available, the experiments have often failed to create violations of the law of large numbers – which is to be expected. The place where the violation of LLN takes place is in a living cell because of the chemical machinery that expends energy to enforce an organization that violates the law of large numbers – i.e. the nucleotide and amino acid synthesis pathways. Sometimes the problem can be over-ridden slightly by catalysts like clay for select (but not all) amino acids, but the result is not persistent because of spontaneous racemization. The problem is orders of magnitute worse for nucleotides.
A robot can hypothetically be programmed to force a system of randomly flipped fair coins to be re-organized to be 100% heads. It’s certainly possible to violate the law of large numbers by creating sufficient machinery to over-ride the tendency of random outcomes.
The molecules of life have an inherent tendency for certain random outcomes outside of a cell. That’s one of the mechanistic reasons Virchow’s principle holds.
There are certain reasons for random outcomes in some contexts, starting with spontaneous isomerization and/or equilibirum concentrations of isomers in solution.
A fair discussion, imho, that is pure science, is exploring the mechanistic reasons Virchow’s principle holds in the present day. How extensible Virchow’s principle is to the question of past abiogenesis is of course a point of sharp debate.
The moral of the story is the 2nd Law of Themrodyanmics is a TERRRIBLE argument in favor of design and/or creationism. Worse than trying to apply CSI.